Instructions

The following file types are supported: swc, dat, asc or json
if you want to use the following software without selecting neurons: 3DspineS, 3DSomaMS, 3DSynapsesSA, Dendrite arborization simulation
to analyze some random neurons in order to understand how NeuroSuites works.
- NeuroMorpho.org: search through the NeuroMorpho.org database to get neurons.
Step 1
Select or not a datasetStep 2
Select an application and analyze the dataList of applications
L-Measure - Extract morphological measurements
This tool allows researchers to extract quantitative morphological measurements from neuronal reconstructions.
Neuronal reconstructions are typically obtained from brightfield or fluorescence microscopy preparations using applications such as Neurolucida, Eutectic, or Neuron_Morpho, or can be synthesized via computational simulations.
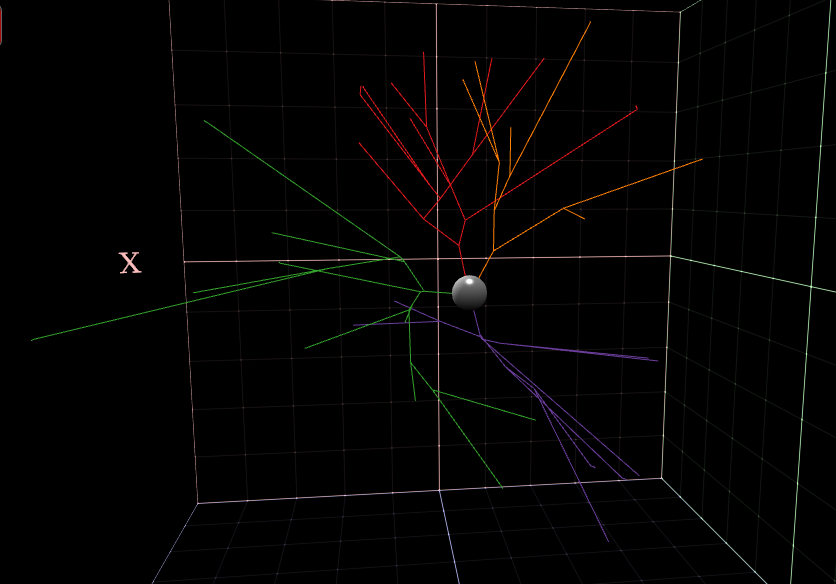
NeuroViewer - 3D Neuron reconstruction
3D Neuron reconstruction visualization package
NeuroSTR was originally created by Luis Rodriguez-Lujan.
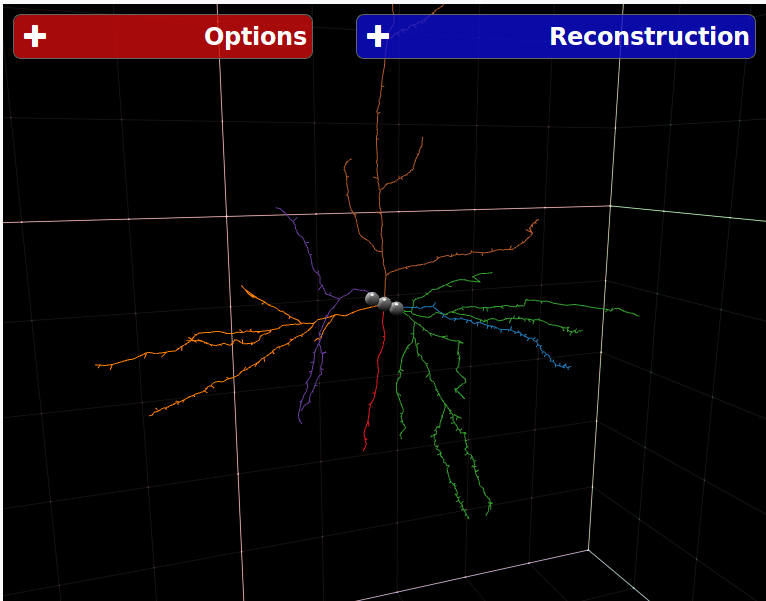
NeuroSTR - Validator, format converter
NeuroSTR is a neuroanatomy toolbox for C++. It reads and processes three-dimensional neuron reconstructions in the most common file formats and offers a huge set of functions and utilities to work with them.
NeuroSTR was originally created by Luis Rodriguez-Lujan and is currently maintained by Bojan Mihaljevic.


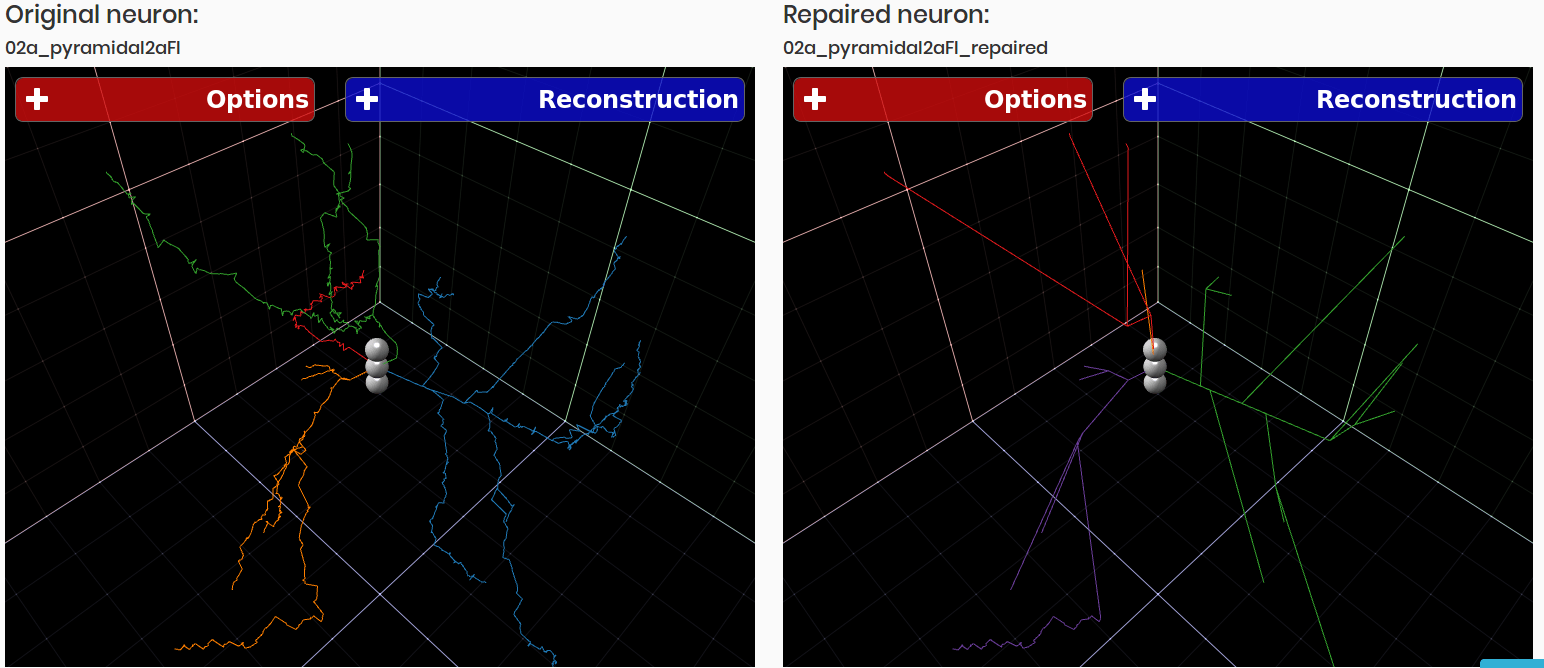
3DBasalRM - Repair cut-points in the basal arborization
Data-driven repairing model that detects cut-points in the basal arborization and then repairs them using a growth model built from complete three-dimensional neuron reconstructions.
As result a neuron in JSON format is returned
3DBasalRM was originally created by Sergio Luengo.
GabaClassifier - Interneuron classifier
Classifies the given interneuron morphology into one of the 7 possible classes.
The model has been trained with layer L2/3 to layer L6 interneurons and thus only interneurons from those layers are allowed as input.
Click here to learn more about the interneuron classes)
GabaClassifier was originally created by Bojan Mihaljevic
Statistics engine
Discrete and continuous data are supported.
Descriptive statistics:univariate, bivariate and multivariate analysis and visualization.
Inferential statistics: confidence intervals, hypothesis testing (one sample t-test, two dependent and independent samples t-test), find fittest distribution.
Interactive plots with Plotly (histograms, probability density functions, box plots, 2D and 3D scatter plots, Chernoff faces, Radar charts, Parallel coordinates, Andrew curves and much more!), custom options, exporting formats, etc.
Everything online.
Check out the Data stats or L-Measure tool in the Morphometric Analyzer to see it in action.

Machine learning
Bayesian Networks: structure and parameters learning for continuous and discrete datasets. Full visualization and inference for continuous BNs.
Probabilistic clustering graphical models: full visualization and inference for continuous models.
Everything online.
Check out the Machine learning section in the Morphometric Analyzer to see it in action.
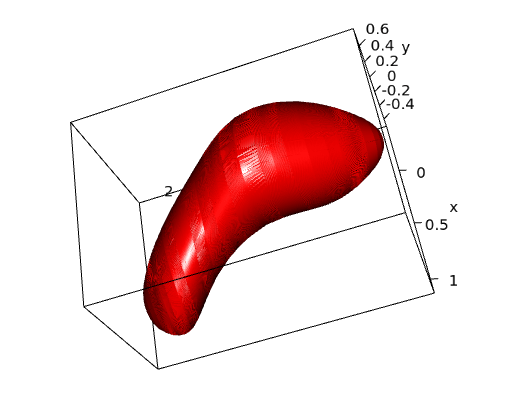
This mathematical approach could provide a useful tool for theoretical predictions on the functional features of human pyramidal neurons based on the morphology of dendritic spines.
This tool was trained with human cortical pyramidal neurons.
3DspineS was originally created by Sergio Luengo.
Luengo-Sanchez, S., Fernaud-Espinosa, I., Bielza, C., Benavides-Piccione, R., Larrañaga, P., & DeFelipe, J. (2018).
3D morphology-based clustering and simulation of human pyramidal cell dendritic spines.
PLOS Computational Biology, 14(6), e1006221. https://doi.org/10.1371/journal.pcbi.1006221
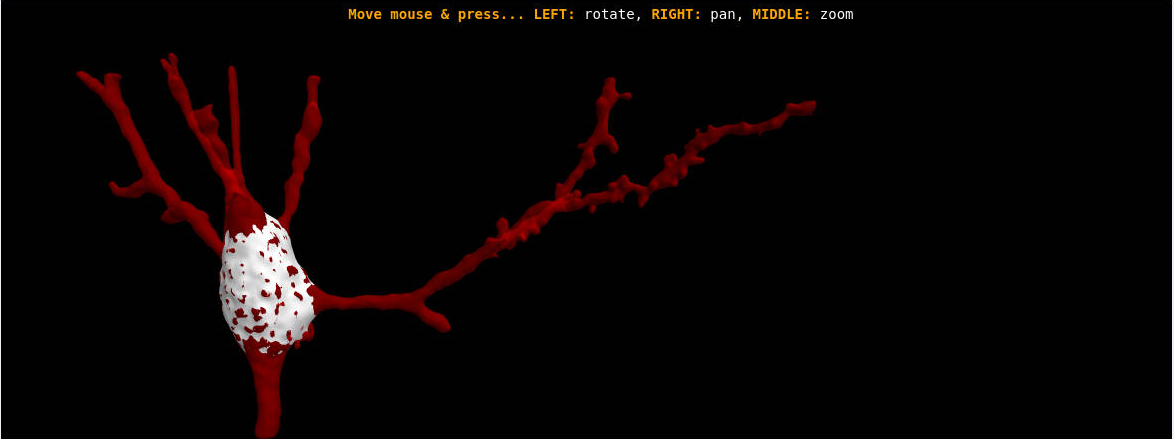
The definition of the soma is fuzzy, as there is no clear line demarcating the soma of the labeled neurons and the origin of the dendrites and axon. Thus, the morphometric analysis of the neuronal soma is highly subjective.
This software provides a mathematical definition and an automatic segmentation method to delimit the neuronal soma. We applied this method to the characterization of pyramidal cells, which are the most abundant neurons in the cerebral cortex. Thus, this software is a means of characterizing pyramidal neurons in order to objectively compare the morphometry of the somata of these neurons in different cortical areas and species.
3DSomaMS was originally created by Sergio Luengo and Luis Rodriguez-Lujan (GUI).
Luengo-Sanchez, S., Bielza, C., Benavides-Piccione, R., Fernaud-Espinosa, I., DeFelipe, J., & Larrañaga, P. (2015).
A univocal definition of the neuronal soma morphology using Gaussian mixture models.
Frontiers in Neuroanatomy, 9, 137. https://doi.org/10.3389/fnana.2015.00137
3DSynapsesSA is a tool designed to process and analyze patterns in the three-dimensional spatial distribution of cortical synapses. It brings a variety of both innovative and well-known techniques from the spatial statistics field.
This tool allows you to:
Process and visualize data from cortical synapses for error checking
Model the spatial distribution of the synapses
Replicate, via simulation, samples of cortical synapses
Compare several indicators obtained from data of different layers
3DSynapsesSA was originally created by Laura Antón-Sánchez and Luis Rodriguez-Lujan (GUI).
Generation of synthetic neurons with soma and dendrites.
Dendrite arborization simulation was originally created by Pablo Fernández González